


You can click here to download the 1st Version of the Protocol for model intercomparison on terrestrial carbon cycle with the traceability framework.
Brief introduction of a traceability framework for Model-intercomparison
Biogeochemical models have been developed to account for more and more processes, making their complex structures difficult to be understood and evaluated. Based on the fundamental properties of terrestrial carbon cycle, we developed a framework which traces modeled ecosystem carbon storage capacity (Xss) to several traceable components. These components are (i) Xss as a product of net primary productivity (NPP) and ecosystem residence time (τE), (ii) baseline carbon residence times (τE’), (iii) environmental scalars (ξ), including temperature and water scalars, and (iv) environmental forcings. The relationships between the four components are organized as:
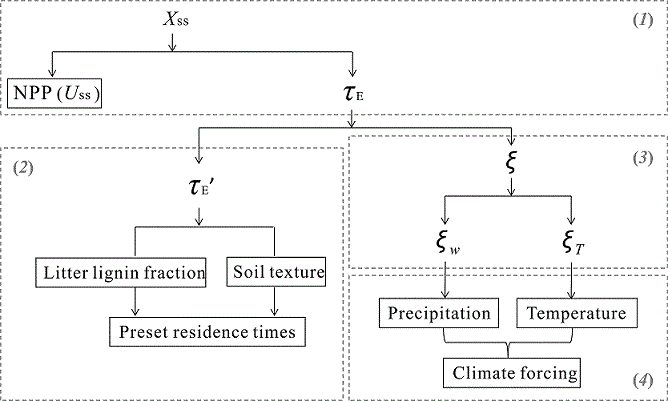
where NPP is net primary productivity, τE is ecosystem carbon residence time, τE' is baseline ecosystem carbon residence time, ξ is environmental scalar on carbon decay rates (ξw and ξT for water and temperature scalar, respectively).
This traceability framework has been successfully applied to the Australian Community Atmosphere Biosphere Land Exchange (CABLE) model to help understand differences in modeled carbon processes among biomes and as influenced by nitrogen processes. This framework is very useful for model intercomparisons on model structures of terrestrial biogeochemistry. More details about this study can be found in our paper on Global Change Biology (Xia et al. 2013).
Required output from the model
1. Develop a flow diagram to represent model structure (box-arrow) as we have done for the CABLE model(this diagram is helpful but not required):
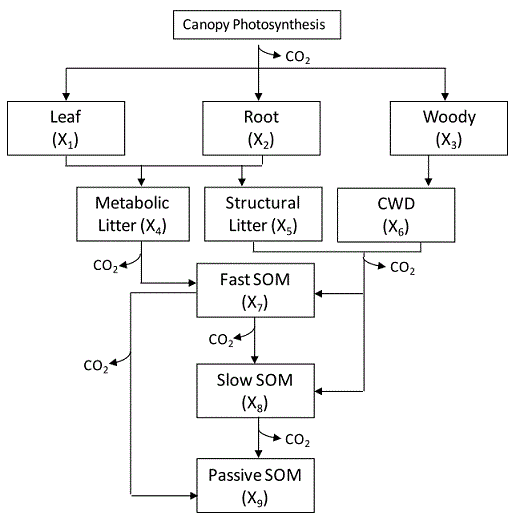
2. Find carbon balance equation for each of the pools with input and output (required);
3. Average NPP at steady state;
4. partitioning coefficient of NPP to each plant pool;
5. Potential mortality rate of each plant pool;
6. Adjusting factor for mortality rate (every scalars effect on mortality);
7. Transfer coefficient of dead plant tissue carbon from each plant pool to each litter pool;
8. Potential decay rate of litter from each litter pool;
9. Adjusting factor for potential decay rate;
10. Carbon transfer from each litter pool to each of soil pool;
11. Potential decomposition rate of soil organic carbon from each of soil pool;
12. Adjusting factors for decomposition rate;
13. Carbon transfer from each of soil pool to other soil pools.
Required variables
In order to make the required outputs more clear, we summarized them into the following table:
Variable |
Associated Component |
Deblahion |
Net primary production (NPP) |
Carbon influx |
|
Allocation of NPP to leaf biomass |
B vector |
dimensionless; |
Allocation of NPP to root biomass |
B vector |
dimensionless; |
Allocation of NPP to woody biomass |
B vector |
dimensionless; |
Transfer coefficient from leaf biomass to metabolic litter |
A matrix |
dimensionless; |
Transfer coefficient from leaf biomass to structural litter |
A matrix |
dimensionless; |
Transfer coefficient from woody biomass to CWD |
A matrix |
dimensionless; As '1' since all woody biomass transfer to CWD. |
Transfer coefficient from metabolic litter to fast SOM |
A matrix |
dimensionless; |
Transfer coefficient from structural litter to fast SOM |
A matrix |
dimensionless; |
Transfer coefficient from structural litter to slow SOM |
A matrix |
dimensionless; |
Transfer coefficient from CWD to fast SOM |
A matrix |
dimensionless; |
Transfer coefficient from CWD to slow SOM |
A matrix |
dimensionless; |
Transfer coefficient from fast SOM to slow SOM |
A matrix |
dimensionless; |
Transfer coefficient from fast SOM to passive SOM |
A matrix |
dimensionless; |
Transfer coefficient from slow SOM to passive SOM |
A matrix |
dimensionless; |
Potential turnover rate of leaf biomass |
C matrix |
year-1; |
Potential turnover rate of root biomass |
C matrix |
year-1; |
Potential turnover rate of woody biomass |
C matrix |
year-1; |
Potential turnover rate of metabolic litter |
C matrix |
year-1; |
Potential turnover rate of structural litter |
C matrix |
year-1; |
Potential turnover rate of CWD |
C matrix |
year-1; |
Potential turnover rate of fast SOM |
C matrix |
year-1; |
Potential turnover rate of slow SOM |
C matrix |
year-1; |
Potential turnover rate of passive SOM |
C matrix |
year-1; |
Water limitation on leaf biomass |
Water scalar (ξw) |
dimensionless; |
Water limitation on root biomass |
Water scalar (ξw) |
dimensionless; as '1' in the CABLE model |
Water limitation on woody biomass |
Water scalar (ξw) |
dimensionless; as '1' in the CABLE model |
Water limitation on decomposition of metabolic litter |
Water scalar (ξw) |
dimensionless; |
Water limitation on decomposition of structural litter |
Water scalar (ξw) |
dimensionless; |
Water limitation on decomposition of fast SOM |
Water scalar (ξw) |
dimensionless; |
Water limitation on decomposition of slow SOM |
Water scalar (ξw) |
dimensionless; |
Water limitation on decomposition of passive SOM |
Water scalar (ξw) |
dimensionless; |
Temperature limitation on leaf biomass |
Tempearture scalar (ξT) |
dimensionless; |
Temperature limitation on root biomass |
Tempearture scalar (ξT) |
dimensionless; as '1' in the CABLE model |
Temperature limitation on woody biomass |
Tempearture scalar (ξT) |
dimensionless; as '1' in the CABLE model |
Temperature limitation on decomposition of metabolic litter |
Tempearture scalar (ξT) |
dimensionless; |
Temperature limitation on decomposition of structural litter |
Tempearture scalar (ξT) |
dimensionless; |
Temperature limitation on decomposition of fast SOM |
Tempearture scalar (ξT) |
dimensionless; |
Temperature limitation on decomposition of slow SOM |
Tempearture scalar (ξT) |
dimensionless; |
Temperature limitation on decomposition of passive SOM |
Tempearture scalar (ξT) |
dimensionless; |
Pool size of leaf |
Steady-state pool size (Xss) |
g C m-2; |
Pool size of root |
Steady-state pool size (Xss) |
g C m-2; |
Pool size of woody biomass |
Steady-state pool size (Xss) |
g C m-2; |
Pool size of metabolic litter |
Steady-state pool size (Xss) |
g C m-2; |
Pool size of structural litter |
Steady-state pool size (Xss) |
g C m-2; |
Pool size of CWD |
Steady-state pool size (Xss) |
g C m-2; |
Pool size of fast SOM |
Steady-state pool size (Xss) |
g C m-2; |
Pool size of slow SOM |
Steady-state pool size (Xss) |
g C m-2; |
Pool size of passive SOM |
Steady-state pool size (Xss) |
g C m-2; |
Annual total precipitation |
Forcing |
mm |
Annual air temperature |
Forcing |
K |
Desired model information
1. Response functions of carbon process to temperature and precipitation (or moisture ), or individual response functions for each pool);
2. Soil texture map;
3. Response functions to link soil texture to soil C processes;
4. Lignin fraction and other associated factors in affecting litter C processes;
5. Vegetation map.
Common guidelines for model runs
Each model can use their own forcings to drive the model, and specific spin-up method to run the model to steady state. Here are some common requirements for the model outputs:
(1) Models outputs should be obtained from the steady state;
(2) No additional simulations are needed for this model intercomparion, but the modelers should find the variables in Table 1 in their models and output them at the end of spin-up simulation.
(3) All the variables in the Table 1 are required as temporal averages from the steady-state run, but daily outputs of all variables are preferred.
Distribution policy
The traceability framework is distributed free to the academic community on the following conditions:
*Works based on the traceability framework leading to scientific publications should cite the paper published by Xia et al. (2013).
Reference
Xia JY, YQ Luo, YP Wang, O Hararuk. Traceable components of terrestrial carbon storage capacity in biogeochemical models. Global Change Biology 19(7):2104–2116.[Download PDF]
